Welcome to this second blog post on genetics/genomics for drug discovery! So far, we are 2 for 2. That is, this is the second week in a row where we have reviewed the literature for interesting journal articles and written a blog on why the study is relevant for drug discovery. I say “we”, because this week I asked for input from our Merck Genetic & Pharmacogenomics (GpGx) team. We received a number of interesting submissions from GpGx team members, as summarized at the end of the blog.
This week’s article uses antisense as therapeutic proof-of-concept in humans for a genetic target…again! This story is reminiscent of last week’s post on APOC3 (see here).
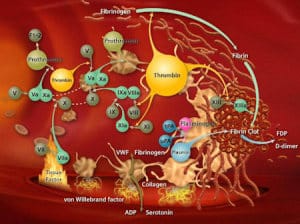
Factor XI Antisense Oligonucleotide for Prevention of Venous Thrombosis, New England Journal of Medicine (December 2014).
Summary of the manuscript: While patients with congenital Factor XI deficiency have a reduced risk of venous thromboembolism (VTE), it is unknown whether therapeutic modulation of Factor XI will prevent venous thromboembolism without increasing the risk of bleeding. In this open-label, parallel-group study, 300 patients who were undergoing elective primary unilateral total knee arthroplasty were randomly assigned to receive one of two doses of FXI-ASO (200 mg or 300 mg) or 40 mg of enoxaparin once daily. The primary efficacy outcome was the incidence of venous thromboembolism. The principal safety outcome was major or clinically relevant non-major bleeding. The primary efficacy outcome occurred in 36 of 134 patients (27%) who received the 200-mg dose of FXI-ASO and in 3 of 71 patients (4%) who received the 300-mg dose of FXI-ASO, as compared with 21 of 69 patients (30%) who received enoxaparin. The 200-mg regimen was noninferior, and the 300-mg regimen was superior, to enoxaparin (P<0.001). Bleeding occurred in 3%, 3%, and 8% of the patients in the three study groups, respectively.
Why this is interesting: Human genetics can be used to estimate both efficacy and safety (see slide #5 in slide deck here). In the case of Factor XI as a therapeutic target, genetic deficiency of Factor XI leads to very mild bleeding liability, much more mild than other Factor deficiencies (e.g. Factor X, see here). The limited data available suggests that perturbation of Factor XI may lead to less bleeding liability than other anti-coagulants. But is it effective? The available population-based data on homozygous carriers of rare F11 mutations that abolish gene function suggest that Factor XI deficiency protects from deep vein thrombosis (see here) and stroke (here). Further, there is genetic support from GWAS that common alleles in Factor XI protect from VTE, lower serum factor XI levels and alter aPTT (see here, here, here).
But genetic data alone only generates therapeutic hypotheses. Ultimately, testing in humans directly is required to test these hypotheses, as there are many reasons why genetic targets may ultimately fail in the clinic (see here). For the first time, this NEJM paper tests this genetic therapeutic hypothesis for VTE, and the answer is a resounding success.
In summary, I conclude that together, these data provide evidence that LOF mutations in the F11 gene protect from a complex trait, VTE, and that therapeutic perturbation of Factor XI recapitulates the human genetic data. If you are keeping track at home, I would add F11 as a gene to the short list of LOF-protective genes, similar to PCSK9, CCR5, APOC3, SLC30A8, IFIH1, CARD9, etc. [Thanks to Joe Maranville and Dermot Reilly for help on this submission.]
Other articles of interest:
Mutations in the deubiquitinase gene USP8 cause Cushing’s disease, Nature Genetics (December 2014). This study builds on a body of evidence published earlier in the year on benign cortisol-producing adrenocortical tumors (see Science commentary here). The significance of the Nature Genetics paper is described in a press release from Tokyo Institute of Tech: The researchers conclude that their results “not only identify the first of so far enigmatic driver mutations in corticotroph adenomas but also elucidate a novel mechanism by which the EGFR pathway is constitutively activated in human tumours.” One could imagine developing inhibitors of USP8 for the treatment of Cushing’s disease. [Thanks to Richard Chen for this submission.]
PD-1 Blockade with Nivolumab in Relapsed or Refractory Hodgkin’s Lymphoma, New England Journal of Medicine (December 2014). Yet another study showing the promise of immuno-oncology. In this ongoing study, 23 patients with relapsed or refractory Hodgkin’s lymphoma that had already been heavily treated received nivolumab (at a dose of 3 mg per kilogram of body weight) every 2 weeks until they had a complete response, tumor progression, or excessive toxic effects. An objective response was reported in 20 patients (87%), including 17% with a complete response and 70% with a partial response; the remaining 3 patients (13%) had stable disease. An interesting component of the study was the assessment of copy number variation at the PDL1-PDL2 locus on chromosome 9p in the subgroup of 10 patients with available tumor samples. In all tumors analyzed by means of FISH, tumor cells had 3 to 15 copies of PDL1 and PDL2 in patterns characterized by amplification, relative copy gain, or polysomy of chromosome 9p.
Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction, Nature (December 2014). After sequencing the exons of APOA5 in 6,721 cases and 6,711 controls, the study identified 46 unique non-synonymous or splice-site SNVs or indel frameshifts with allele frequency <1%. A burden of rare mutations in APOA5 explains about 0.14% of the total variance for MI and roughly 0.28% of the heritability. After sequencing the exons of LDLR in 4,703 cases and 5,090 controls, the study identified 156 unique non-synonymous, splice-site SNVs and indel frameshifts with allele frequency <1%. A burden of rare mutations in LDLR explains about 0.24% of the total variance for MI and roughly 0.48% of the heritability. Using these rare variant signals as a guide, the study estimated sample sizes that will be required to make similar discoveries: >10,000 exomes are required to achieve 80% statistical power at an exome-wide level of statistical significance. In addition to these two genes, recent evidence has connected MI risk with variants at two genes functionally related to APOA5, namely lipoprotein lipase (LPL) and apolipoprotein C-III (APOC3). Combined, these observations suggest that disordered metabolism of LDL cholesterol and triglyceride-rich lipoproteins contribute to MI risk. [Thanks to Dermot Reilly, Heiko Runz and Sergey Lezhnin for this submission. Dermot is a co-author on the manuscript.]
Complex host genetics influence the microbiome in inflammatory bowel disease, Genome Medicine (December 2014). This manuscript finds a novel mechanism of action for the well-known association of NOD2 risk variants and inflammatory diseases of the gut: influencing composition of gut microbiota. As the authors point out, microbial DNA is considered a secondary genome with potentially heritable signals. The proposed dysbiosis mechanism of action both explains NOD2 association with multiple diseases and the noisiness of the association, clarifying the previously observed human genetic signal. To accomplish this clarification, the authors carefully integrate multiple data types, segregating environmental and artifactual causes of variation, such as biopsy characteristics and recent treatment, from human genetic drivers. The dysbiosis mechanism of NOD2 action suggests novel means of therapeutic intervention that would not have made sense without following up on the genetic lead. [Thanks to Julja Burchard for this submission.]
Conserved Higher-Order Chromatin Regulates NMDA Receptor Gene Expression and Cognition, Neuron (November 2014). This paper by Bharadwaj et al focused on non-coding sequences that control higher-order chromatin structure around the NMDA glutamate receptor GRIN2B. Using chromosome conformation capture, authors analyzed ~800kb noncoding sequence encompassing GRIN2B and identified two long-range chromosomal loop formations that are specific to brain, characterized by peaks in acetylated histone H3, associated with the expression of GRIN2B, and regulated by synaptic activity. The loops serve to carry positive transcriptional regulators bound to intronic or intergenic sequences into close proximity to the transcription start site of GRIN2B. The authors interrogated the Psychiatric Genome Consortium 2 and identified a risk polymorphism in the distal loop that is associated with decreased GRIN2B expression in the context of schizophrenia. The risk allele was further associated with cognition – healthy subjects from the LOGOS cohort underwent neuropsychological testing, and those with the T-allele at rs117578877 performed less well in spatial memory tasks and more frequently displayed personality traits associated with schizophrenia spectrum disorders. Further, the authors employed CRISPR to engineer targeted mutations in proximity to rs117578877, and found that these mutations (449kb from the transcription start site) were associated with decreased GRIN2B expression in transfected cells. [Thanks to Andi Webber for this submission.]
Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia, Nature Medicine (December 2014). This paper shows a very comprehensive functional validation of a human genetic finding. The authors engineer a genetically relevant mouse model of Frontotemporal Dementia with an inducible forebrain transgene expressing a mutant CHMP2B protein that is truncated at the carboxy terminus (CHMP2BIntron5), which is thought to result in toxicity. Mice expressing the mutant protein show an age-related sociability phenotype. At the molecular level, the authors show that the phenotype is linked to altered postsynaptic AMPA receptor levels (up-regulation) and function. They hypothesize a role for a microRNA in this up-regulation (a bit of a leap in this case but certainly could be based on RNASeq data) and show the miR-124 is down-regulated upon transgene expression and that some of the AMPA receptors, thought to be targets of miR-124 are thereby up-regulated. They demonstrate relevance to human disease by showing similar differences in miR-124 and AMPAR expression in subjects with FTD. For additional functional validation they studied miR-124 and AMPARs in iPSC lines derived from three FTD patients. When they derived neurons from these cells, levels were the same for miR-124 and AMPARs in early neurons but changed versus control as the neurons aged in the dish. For proof of concept for the miR-124 role, they used AAV to express a mir-124 mimic in the transgenic mouse model and showed that it could reverse the phenotype. They also tested the role of the AMPARs by constructing lentiviral vectors encoding an shRNA directed against one of the mouse receptors and showing that its knockdown partially rescued the phenotype. The therapeutic path here is questionable as microRNA therapies remain problematic; however, there are some obvious ways to think about creating a phenotypic screen to either down-regulate the aberrant AMPARs or up-regulate miR-124 in aged FTD patient IPSC-derived neurons. [Thanks to Michele Cleary for this submission.]