This week’s theme is genes to function for drug screens…with a macabre theme of zombies! As more genes are discovered through GWAS and large-scale sequencing in humans, there is a pressing need to understand function. There are at least two steps: (1) fine-mapping the most likely causal genes and causal variants; and (2) functional interrogation of causal genes and causal variants to move towards a better understanding of causal human biology for drug screens (“from genes to screens”).
Genome-editing represents one very powerful tool, and the latest article from the laboratory of Feng Zhang at the Broad Institute takes genome-editing to a new level (see Genetic Engineering & Biotechnology News commentary here). They engineer the dead!
Genome-scale gene activation by an engineered CRISPR-Cas9 complex, Nature (December 2014).
Since its introduction in late 2012, the CRISPR-Cas9 gene-editing technology has revolutionized the ways scientists can apply to interrogate gene functions. Using a catalytically inactive Cas9 protein (dead Cas9, dCas9) tethered to an engineered single-guide RNA (sgRNA) molecule, the authors demonstrated the ability to conduct robust gain-of-function genetic screens through programmable, targeted gene activation.
Earlier this year, the laboratories of Stanley Qi, Jonathan Weissman and others \ reported the use of dCas9 conjugated with a transcriptional activator for gene activation (see Cell paper here).…
Read full article...

Welcome to this second blog post on genetics/genomics for drug discovery! So far, we are 2 for 2. That is, this is the second week in a row where we have reviewed the literature for interesting journal articles and written a blog on why the study is relevant for drug discovery. I say “we”, because this week I asked for input from our Merck Genetic & Pharmacogenomics (GpGx) team. We received a number of interesting submissions from GpGx team members, as summarized at the end of the blog.
This week’s article uses antisense as therapeutic proof-of-concept in humans for a genetic target…again! This story is reminiscent of last week’s post on APOC3 (see here).
Factor XI Antisense Oligonucleotide for Prevention of Venous Thrombosis, New England Journal of Medicine (December 2014).
Summary of the manuscript: While patients with congenital Factor XI deficiency have a reduced risk of venous thromboembolism (VTE), it is unknown whether therapeutic modulation of Factor XI will prevent venous thromboembolism without increasing the risk of bleeding. In this open-label, parallel-group study, 300 patients who were undergoing elective primary unilateral total knee arthroplasty were randomly assigned to receive one of two doses of FXI-ASO (200 mg or 300 mg) or 40 mg of enoxaparin once daily.…
Read full article...
There are an increasing number of very interesting published studies around genetics / genomics and drug discovery. Just last week, there were a series of articles in Nature on predictors of response to anti-PD1 therapy. (Disclaimer: I work for Merck, which markets an anti-PD1 drug.) In this new blog series, I try and pick at least one paper that highlights some of the key principles of genetics/genomics and drug discovery. This is week #1…hopefully I will be able to do this routinely!
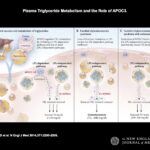
This week, I have selected an article in NEJM by Gaudet et al: Targeting APOC3 in the Familial Chylomicronemia Syndrome. As described in the introduction: “The familial chylomicronemia syndrome is a rare autosomal recessive disease characterized by the buildup in the blood of fat particles called chylomicrons (chylomicronemia), severe hypertriglyceridemia, and the caused by mutations in the gene encoding LPL or, less frequently, by mutations in genes encoding other proteins necessary for LPL function. Patients with this syndrome have plasma triglyceride levels ranging from 10 to 100 times the normal value (1500 to 15,000 mg per deciliter [17 to 170 mmol per liter]), eruptive xanthomas, arthralgias, neurologic symptoms, lipemia retinalis, and hepatosplenomega without pancreatitis, that interfere with normal life and result in frequent hospitalizations.”…
Read full article...




