At the Harvard-Partners Personalized Medicine Conference last week I participated in a panel discussion on complex traits. When asked about where personalized medicine for complex traits will be in the future, I answered that I envision two major categories for personalized therapies.
(1)Development of drugs based on genetic targets will lead to personalized medicine; and
(2)Large effect size variants will be detected in clinical trials or in post-approval studies and will lead to personalized medicine.
This answer, I said, was based in part on current categories of FDA pharmacogenetic labels and in part on how I see new drug discovery occurring in the future. But did the current FDA labels really support this view?
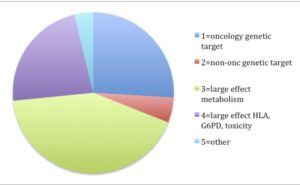
The answer is “yes”. In reviewing the 158 FDA labels (Excel spreadsheet here), my crude analysis found that 31% of labels fall into the “genetic target” category (most from oncology – 26% of total) and 65% fall into the “large effect” category (most from drug metabolism [42% of total], HLA or G6PD [15% of total]).
A subtle but important point is that I predict that category #2 (PGx markers for non-oncology “genetic targets”) will grow in the future. In other words, development of non-oncology drugs will riff-off the success of drugs developed based on somatic cell genetics in oncology. The drugs in that category today include maraviroc (based on CCR5 mutations), ivacaftor (CFTR mutations), warfarin (VKORC1 mutations), and statins (based on LDLR mutations). [One could argue whether statins should fall into this category, as the target of statins is HMGCR not LDLR per se…but for now, please accept this liberal classification.]
Increasingly, I predict we will use human genetics to select novel drug targets at the front end of the drug discovery engine (here). Based on this model, human genetics will not only reveal novel targets for complex traits, but also help select biomarkers for target engagement (i.e., to make sure the drug is doing what it should be doing) and potentially for patient stratification (i.e., personalized medicine).
Conceptually this idea makes sense: if drug targets are selected based on human genetics, then these same genetic findings can be used to find biomarkers that uncover biology that differentiates between human carriers vs non-carriers of the same mutations. These biomarkers, in theory, can be used to find patients who have molecular subsets of disease most amenable to treatment by a genetic-based molecular mechanism of action. Most of the time, I predict that the biomarkers will NOT be genetic variants themselves, but rather protein or mRNA biomarkers based on genetic findings – not too dissimilar for the way in which the protein periostin is used to differentiate asthma patients for anti-IL13 therapy (here).
Occasionally, the same genetic variants used to select the targets may be used in patient stratification. This is the case with maraviroc (based on CCR5 mutations) and ivacaftor (CFTR mutations). It is worth noting that this is unlikely to be the case with PCSK9 inhibitors, which, if approved, will be for LDL lowering regardless of PCSK9 genotype.
A recent manuscript published in Science Translational Medicine (here) supports this theme of genes-to-drugs-to-biomarkers. A diabetes-associated genetic variant in ADRA2A causes alpha-2A-adrenergic receptor overexpression and impaired insulin secretion. An antagonist of this receptor, yohimbine, reversed the defect observed in patients carrying the risk genotype. The authors conclude: “The findings show that knowledge of genetic risk variants can be used to guide therapeutic interventions that directly target the underlying pathophysiology and demonstrate the potential of individualized genotype-specific treatment of type 2 diabetes.”
Based on this view, I predict that pharmacogenetics focused on two distinct activities (genetic targets and large effect size variants) will have different time horizons and scope of impact: (1) If genetics is used to find new drug targets, then the application of routine personalized medicine is many years away, as it takes approximately 10-15 years to develop drugs once a target is identified. While farther away, the scope of impact could be much greater – potentially most drugs will have some companion diagnostic test based on biology derived from genetics (e.g., proteins, expression signatures) or based on the genetic variants themselves (e.g., what we consider today as traditional pharmacogenetics). (2) If large effect variants is the major driver of personalized medicine, then I predict that personalized medicine for complex traits will continue at its current pace and scope – which is slow and steady, but without a major impact on most drugs used in clinical practice.