[I am an employee of Celgene. All views expressed here are my own.]
At the 2018 Annual Atlas Ventures Retreat (AVR), I participated in a panel on Digital Health (along with David Schenkhein, John Reed, Scott Brun). The panel discussion was led by Michael Ringel, who also provide an excellent introduction to Digital Health (his slides here). While there are many aspects to digital health, we focused on the application to drug discovery and development. In this blog, the main point I want to emphasize is that I believe that the digital health tipping point will occur when products that benefit patients (e.g., therapeutics) facilitate the integration of digital health initiatives that currently reside in silos.
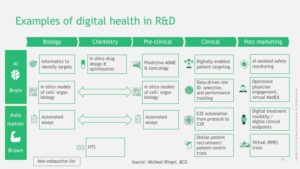
What is digital health in relation to drug discovery & development? There are many different definitions with many different components, and this, in essence, is part of the challenge (see Figure below). In early discovery biology, digital health represents various data types (e.g., human genetics, ‘omics data, cell models) and analytical methods (e.g., simple regression, machine learning, artificial intelligence). In late discovery biology, digital health includes sophisticated analytical methods for in silico drug design and organoid models to recapitulate the human system for pre-clinical testing. In clinical development, digital health encompasses wearables and mobile devices to track patient outcomes in trials. And digital health resides in the messy and unstructured “real world”: AI-assisted clinical data interpretation (e.g., radiology, pathology), virtual cohorts, and electronic health records.
Currently, most of these digital health applications reside in silos. Human genetics and genomics is largely based in research environments. Wearables and mobile devices are primarily used by consumers, although are increasingly adopted in clinical trials. Electronic Health Records (EHRs) are restricted to clinical care. Virtual clinical trial cohorts are for small subsets of patients.
What are the activities that will break down these silos? Below I list three predictions that are unified by a real-world / human genetics thread: human genetic discoveries derived from biobanks will be used to start a drug discovery journey; biomarker and clinical endpoints derived from real-world data, and validated through Mendelian randomization, will be used to test proof-of-concept in early development; and virtual cohorts selected based on modeling of genetics and standard-of-care therapies will be used in late development clinical trials. In other words, data used to validate a human genetic target will also be carried forward into pre-clinical testing, early proof-of-concept trials, and registration clinical trials.
1. Discoveries made from real-world digital health data will start the drug discovery journey. Most human genetic discoveries are made from case-control cohorts collected outside of real-world data. However, the rise of biobanks is changing this (see announcement on a 5 million genome project in the UK here). Increasingly, discoveries will be made and/or implemented within these biobanks. For example, if a genetic variant is linked to clinical outcomes, and these outcomes are surrogates for therapeutic efficacy and on-target toxicity, then this is the starting point for a drug discovery journey. I have used TYK2 and psoriasis as an example (see the YouTube video here from my recent presentation at the American Society of Human Genetics Annual Meeting).
2. The same real-world data used to select a genetic target will be used to establish clinical proof-of-concept in pre-specific patient populations. Mendelian randomization is a powerful tool to establish causality between a biomarker and clinical outcome. If a genetic target is linked to a biomarker using real-world data, and a therapeutic modality is matched to the molecular mechanism of action, then these same biomarkers can be used to demonstrate clinical proof-of-concept. A simple example is PCSK9 and LDL cholesterol. However, as biobanks expand, and as PheWAS are conducted using real-world data, I predict that many new biomarkers – and in some instances unexpected biomarkers – will also serve as readouts in clinical trials. Moreover, because real-world data includes outcomes from patients on approved therapies, and because human genetics can be used to model networks of approved vs novel therapies, it should be possible to pre-specify patients most likely to benefit from a novel therapeutic (see blog, Precision Medicine by Design for examples).
3. Virtual cohorts will be used to demonstrate clinical efficacy relative to standard-of-care…and ultimately for registrational trials. Late-stage clinical trials contribute disproportionately to R&D expense. One reason is the challenge of patient recruitment: some patients don’t want to be randomized to a placebo arm, which slows overall enrollment. One approach to overcome this challenge is to use real-world “virtual cohorts” as the comparison arm. Of course, such virtual cohorts carry their own baggage, including the concern that the comparison cohort is not an appropriate match to the clinical trial cohort. However, if a drug target is based on human genetics, and human genetics plus real-world was used to model treatment response, then it should be possible to more accurately select comparison cohorts and minimize bias. Clearly a lot of work needs to be done to establish confidence in this model…but it seems possible. Dr. Scott Gottlieb’s recent Sunday Tweetorial (here) on real-world data for regulatory approval provides evidence in support of this future state.
Again, the main point is that it is the integration of digital health applications across silos that will ultimately lead to the incorporation of digital health in drug R&D. I have used human genetics as one way to integrate across silos, but there are many other examples of technology breakthroughs that will remove barriers. In his AVR introduction, Michael used the development of drugs to cure HCV to emphasize this point: breakthroughs happen when all the “hurdles” comes down (see slide #4). He also cited the deluge of digital data (slide #5) and advances in computing (see slide #6) as drivers of change.
To close, I want to highlight recent digital health news stories. The purpose of these links is to reinforce my argument that digital health is happening, which is probably not a surprise to most readers. The links also point to challenges of implementing digital health – including a convincing essay by David Shaywitz on dangers of metric, measurement, and religious managerialism.
- Digital health is happening:
- Keystone Symposium on Digital Health (here)
- Apple and VA health system (here)
- Google Hires Geisinger CEO Dr. David Feinberg (here, here)
- Amazon HQ announced, and with it predictions of “Amazon Health Care” (here)
- A roadmap by Vijay Pande of how to engineer biology (here)
- Deloitte’s annual “Unlocking R&D productivity”, which emphasizes digital health (here)
- GE files paperwork to spin out health-care unit (here)
- Apple watch as personal ECG (here)
- Daphne Koller on “Human biology through the lens of data” (here)
- STAT opinion piece on Google and digital health (here)
- Digital health market is frothy (here)
- But it is not easy:
- Dangers of metric, measurement, and religious managerialism (here)
- Stories of failed digital health start-ups (here)
- Challenges of using the tech’s “move fast and break things” model for digital health companies (here)
- Challenges in the development of glucose sensing contact lens (here)
- Challenges of implementing artificial intelligence (AI) in drug discovery (here)
- A poignant New Yorker essay by Atul Gawande on why doctors hate electronic health records (here)