I attended the Mendelian randomization meeting in Bristol, UK this past week (link to the program’s oral abstracts here). The meeting was timed with the release of a number of articles in the International Journal of Epidemiology (current issue here, Volume 44, No. 2 April 2015 TOC here). This blog is a brief synopsis of the meeting – with a focus on human genetics and drug discovery. The blog includes links to several slide decks, as well as references to several published reviews and studies.
[Disclaimer: I am a Merck/MSD employee. The opinions I am expressing are my own and do not necessarily represent the position of my employer.]
Several speakers, including Lon Cardon from GSK, gave overview talks on how Mendelian randomization can be applied to pharmaceutical development. In my overview, I described important guiding principles for successful drug discovery (link to my slides here), and how Mendelian randomization (MR) is applied within this framework. In particular, I emphasized the role of establishing causality in the human system: MR is a powerful tool to pick targets by estimating safety and efficacy (i.e., genotype-phenotype dose-response curves) at the time of target identification and validation; MR is effective at picking biomarkers for target modulation; and MR provides quantitative modeling of clinical proof-of-concept (POC) studies.
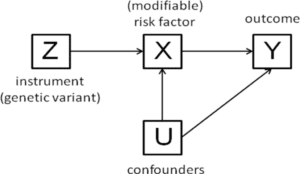
MR requires tools, or instruments, that link human genetic variation to intermediate phenotypes (e.g., biomarkers, cellular assays) and to relevant clinical outcomes (e.g., registration endpoints for clinical trials). Here are a few examples of important tools, including methodological approaches, that were described at the meeting:
– A common theme was the potential for false positives due to pleiotropic instruments, especially in studies that use genetic risk scores built from a large number of variants. These scores often include variants with unclear biological functions, making it difficult to assess the key MR assumption that the only possible path from genotype to outcome is through the intermediate (i.e., the exclusion restriction). Jack Bowden described how MR Egger (or as the cool kids say, “Mister Egger”) regression can be adapted to test for bias from pleiotropy. A manuscript describing the approach can be found in the Int J Epidemiol study (see here). Briefly, genetic effects on outcome are regressed on genetic effects on the intermediate across a set of variants. The slope gives an estimate of the effect of the intermediate on the outcome, and the intercept gives an estimate of the consistent bias across variants (e.g., due to pleiotropic effects). To do so, this method assumes that the strength of pleiotropic effects is not correlated with the strength of the instruments.
– Reproducible genetic associations are a critical first step in any MR study. Further, the greater the number of associated variants, the great the power in MR studies that use aggregate SNP scores. Jeff Barrett, from the Centre for Therapeutic Target Validation (CTTV), described how to fine-map and dissect the biology of GWAS hits, using inflammatory bowel disease (IBD) as an example. To date, more than 200 IBD loci have been identified (unpublished), and Jeff’s team has been able to use new statistical methodologies and genomic datasets to fine-map the most likely causal variant for many of these loci.
– A two-sample MR study design establishes relationships between genetic variants and intermediate phenotypes in one dataset, and then applies the results to another dataset that contains genetic data and clinical traits (see review here). Philip Haycock described a systematic two-sample approach to appraise the causal relevance of telomere length for risk of 81 chronic diseases and 656 non-disease traits, corresponding to 369,184 cases and 864,564 controls. Overall, there was little evidence of association with 652 non-disease traits (except for HDL cholesterol and cell haemoglobin levels), suggesting limited scope for confounding / pleiotropy, helping to recapitulate Mendelian randomization assumptions. Longer telomeres were associated with increased risk of several but not all cancers. There was limited evidence of association with non-cancer diseases (except for coronary heart disease, celiac disease and lung diseases, for which longer telomeres were protective). The majority of results persisted in sensitivity analyses that made allowance for pleiotropy and were to similar to findings based on prospective observational studies.
– Mediation analysis is an approach to deconvolute components of a biological pathway. Maria Glymour applied this method to understand the relationship among components of genetic factors that influence body mass index (appetite, adiposity, cardiopulmonary fitness, “other”) and risk of Alzheimer’s disease. As she said in her talk, the results are “tantalizing but inconclusive”. A related study, recently published in PLoS Medicine by Robert Scott and colleagues, can be found here. A published review on mediation analysis by George Davey Smith can be found here, along with other published examples here and here.
– Brian Ference, from Wayne State University School of Medicine, described a 2×2 factorial method (see link to his recent JACC paper here). He provided examples of how this approach can be used to establish causality for two variables (e.g., two drugs, drug x pharmacogenetic effect, drug x other risk factors).
– Mechanistic studies are essential to interpret results from MR studies. Daniel Freitag described how functional studies of IL6R are used to differentiate between the “classical” and “trans” signaling of membrane-bound and soluble IL6R, respectively.
– Unbiased pleiotropy scans, or Phenome-wide association studies (PheWAS) represent a powerful approach to explore unexpected genotype-phenotype correlations. Zheng-Ming CHEN applied PheWAS to the China Kadoorie Biobank (CKB), a large prospective study of >500,000 individuals from 10 geographic regions in urban and rural China. While it is early days for CKB and other population-based biobanks, these resources will allow for implementation of two-sample MR at an unprecedented scale.
There were several examples of MR in action. Most examples were from the cardiometabolic area. To highlight a few:
– Aroon Hingorani described how MR does not support C-reactive protein (CRP) as a therapeutic target in cardiovascular disease (CVD). However, he described how another inflammatory target, IL6R, may be efficacious in preventing CVD. His slides, which include references to many published studies in this area (e.g., genetic variant V279F in PLA2G7 gene, biomarker LP-PLA2 and CVD), can be found here (link coming soon!).
– Børge Nordestgaard, from the University of Copehagen, also discussed examples in CVD. He described how MR has convincingly established causal relationship between some serum biomarkers and CVD risk [remnant cholesterol, LDL, Lp(a)], but not for others (CRP, HDL). His slides also have many good references for those interested in this topic (link coming soon!).
– Dan Rader described a more complex relationship between HDL and CVD risk. He presented convincing published data that genetic variation in the endothelial lipase gene (LIPG Asn396Ser) is associated with HDL levels, but not associated with CVD risk (Lancet manuscript here). However, he also proposed an “HDL flux hypothesis”: genetic variants that promote efflux of HDL cholesterol from cells, or reverse cholesterol transport, could be beneficial in lowering risk of CVD. A recent Lancet publication by his lab supports this hypothesis (here). Dan’s slides from the meeting can be found here.
– Robert Scott, from the MRC Epidemiology Unit in Cambridge, described MR evidence to support GLP1R agonism as therapeutic approach in type 2 diabetes (see here for Nature Communications manuscript reporting association of the GLP1R variant with fasting glucose). What was quite interesting was how the same GLP1R variant (Ala316Thr; rs10305492) associated with diabetes can be used to predict safety signals such as coronary heart disease (CHD). As stated in his abstract: “the minor allele, which mimicked the effect of GLP1R-agonist therapy, was also associated with lower risk of CHD (OR=0.93[0.87,0.98]; p=0.009).”
Finally, multiple hallway conversations reinforced a few key themes about the future state of MR studies.
– First, power is an issue for most MR studies. Additional large-scale genetic studies are needed to identify many more disease- and trait-associated variants as a starting point for MR studies. For rare variants, many agreed that tens of thousands of patients will be needed to be sequenced to find novel associations. Ideally – at least for the application to the biopharmaceutical industry – these studies should focus on traits that are most relevant for drug discovery.
– Second, better access to datasets is required, especially for a two-sample study design. A limiting factor is that many genetic results are sequestered within supplementary table from publications, making it difficult to perform two-sample MR studies. Increasingly it is interesting to apply MR approaches to assess safety signals, as described by multiple speakers during the conference. These datasets need to be more readily accessible to the research community.
– Third, additional genetic studies are needed for relevant intermediate phenotypes such as serum proteomics or metabolomics, or even more complex biological phenotypes such as cholesterol efflux or telomere length. These datasets will provide the necessary link between a biological target and disease outcomes that are used for registration in clinical trials.
– Fourth, continued methodological development is necessary to establish statistically robust causal relationships. The role of combined genetic risk scores (GRS) vs. individual genes was discussed in multiple settings. Methods such as MR Egger are needed to control for bias. As connections are established, mediation analysis will be important to deconvolute the causal components of genetic pathways. I am particularly interested in quantitative modeling of both efficacy and safety to guide clinical proof-of-concept studies. Such quantitative methods will allow modeling of an “allelic series” model for new drug targets.
– And finally, high-throughput functional studies of associated variants are needed to interpret mechanism (e.g., gain-of-function or loss-of-function). Without knowing direction of effect or mechanism of target perturbation, it is difficult to use MR studies to guide drug development.
To conclude the meeting, George Davey Smith (or as one speaker referred to him, “Big G”) highlighted his take-away themes. He reminded us that MR studies have only been around for about 10-years, although the concept dates back nearly 30 years. Moreover, advanced tools are only now available. He also asked us to consider alternative methods for establishing causal relationships in humans. While MR certainly has limitations, it increasingly represents a potent approach to causal human biology in drug discovery.